Leslie Sieburth
Professor of Biological Sciences
Plant Developmental Genetics

Molecular Biology Program
Education
B.S. Humbolt State University
Ph.D. University of Georgia
Research
My lab focuses on mRNA decay and long-distance signaling pathways using the plant model system Arabidopsis. Specifically, we are studying the roles of P-bodies and mRNA decapping for control of gene expression (RNA level and translational regulation), and a novel signaling pathway used to coordinate developmental processes over long distances.
RNA Decay and P-Body Functions
Analyses of developmental mutants led to identification of mutants with defects in the mRNA decapping enzyme (DCP2/TDT), and mutants with defects in the P-body scaffold protein (VCS/Ge-1/HEDLS). The biochemical reactions leading to removal of an mRNA's 5' cap occur within small cytoplasmic foci called P-Bodies (see figure). The traditional view holds that mRNA decapping and 5' to 3' decay is a bulk pathway that is largely redundant with exosome-mediated 3' to 5' decay. However, our work in Arabidopsis indicates that each of these pathways has specific mRNA substrates (Goeres et al., 2007). These findings lay the groundwork for identifying the determinants that specify an mRNA for a particular pathway.
In plants, microRNAs (miRNAs) have largely been assigned roles in cleavage of their target mRNA, although a few examples of miRNA-directed translational arrest have been documents. We have recently shown that miRNA-directed translational arrest is a wide-spread phenomenon in plants, and that it requires P-bodies (Broderson et al., 2008).
We have also uncovered natural suppressors of mRNA decapping mutants (Goeres et al. 2007). A tremendous resource in Arabidopsis is a large collection natural accession with considerable genetic variation. We find that mRNA decapping mutants show different phenotypes in specific accessions, and we are exploiting this resource to identify the suppressors. We have identified at least two suppressors genetically, and these suppressor function at the RNA level to modify RNA accumulation profiles.
Long-distance Signaling
Research in our lab has uncovered a novel long-distance signaling pathway that uses a novel carotenoid-derived signal (possibly something like retinoic acid). This pathway was identified by characterization of a mutant, bypass1, which over-produces the signal. BYPASS1 encodes a protein of unknown function, and with no functionally characterized domains, but genetically it acts as a negative regulator. How the signal modifies normal development is unknown, but microarray analyses show that the signal is sufficient to drastically reprogram transcription in tissues distant from the signal. Current work in the lab is focused on identifying the signal using biochemical and metabolomic approaches, identifying targets of the signal using molecular, genetic, and cell-biology approaches, and determining how BPS1 functions in the cell. We believe these efforts will allow us to assign the bps1 mobile signal as a new plant hormone.
Developmental Biology
Both the RNA decay and the bps1 long-distance signaling mutants affect normal leaf development. Our long-term goal is to understand how these processes, and other pathways, function in specifying normal development of leaves.
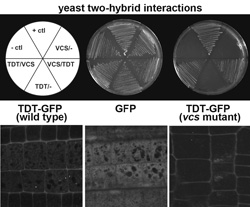
P-body components TDT and VCS interaction and P-Body Formation. Top: The TDT/DCP2 and VCS proteins interact in yeast. Bottom: TDT/DCP2 localizes to discrete cytoplasmic foci, and this localization requires VCS.
References
-
Sorenson, R.S., Deshotel, M.J., Johnson, K., Adler, F.R., and Sieburth, L.E. 2018. Arabidopsis mRNA decay landscape arises from specialized RNA decay substrates, decapping-mediated feedback, and redundancy. Proc. Natl. Acad. Sci. USA. DOI: 10.1073/pnas.1712312115
-
Lee, D.H., Parrott, D.L., Adhikari, E., Fraser, N., and Sieburth, L.E. The mobile bypass Signal Arrests Shoot Growth by disrupting SAM Maintenance, Cytokinin Signaling, and WUS Expression. 2016. Plant Physiology, 171: 2178-2190.
-
Carlos Perea-Resa, Cristian Carrasco-López1, Rafael Catalá, Veronika Turečková, Ondrej Novak, Weiping Zhang, Leslie Sieburth, José Manuel Jiménez-Gómez and Julio Salinas. 2016. The Lsm1-7 Complex Controls Plant Adaptation to Adverse Environmental Conditions By Promoting Selective mRNA Decapping. Plant Cell. 28: 505 – 520.
-
Roux, M.E., Rasmussen, M.W., , Palma, K., Lolle, S., Regué, AM., Bethke, G., Glazebrook, J., Zhang, W., Sieburth, L, Larsen, M.R., Mundy, J., and Petersen, M. 2015. The mRNA decay factor PAT1 functions in a pathway including MAP kinase 4 and immune receptor SUMM2. EMBO J. 34: 593-608.
-
Adhikari, E., Lee, D.-K., Giavalisco, P., and Sieburth, L.E. 2013. Long-distance signaling in bypass1 mutants: bioassay development reveals the bps signal to be a metabolite. Molecular Plant. 6 (1): 164-173.
-
Lee, D-K, and Sieburth, L.E. 2012. Article Addendum: The bps signal: Embryonic arrest from an auxin-independent mechanism in bypass triple mutants. Plant Signaling & Behavior. 7:698-700.
-
Lee, Dong-Keun, Van Norman, J. M, Murphy, C., Adhikari, E., Reed, J. W., Sieburth, L. E. 2012. In the absence of BYPASS1-related gene function, the bps signal disrupts embryogenesis by an auxin-independent mechanism. Development 139: 805-815.
-
Van Norman JM, Murphy C, Sieburth LE (2011) BYPASS1: synthesis of the mobile root-derived signal requires active root growth and arrests early leaf development. BMC Plant Biol 11:28
-
Zhang W, Murphy C, Sieburth LE (2010) Conserved RNaseII domain protein functions in cytoplasmic mRNA decay and suppresses Arabidopsis decapping mutant phenotypes. Proc Natl Acad Sci USA 107:1598
-
Lee DL, Sieburth LE (2010) Plasmodesmata Formation: poking holes in walls with ise. Curr Biol 20:R488-90
-
Belostsky D, Sieburth LE (2009) Kill the messenger: mRNA decay and plant development. Curr Opinion Plant Biol 12: 96-102
-
Brodersen P, Sakvarelidze-Achard L, Bruun-Rasmussen M, Dunoyer P, Yamamoto Y, Sieburth L, Voinnet O (2008) Widespread translational inhibition by plant miRNAs and siRNAs. Science 320:1185-1190
-
Brodersen P, Sakvarelidze-Achard L, Bruun-Rasmussen M, Dunoyer P, Yamamoto Y, Sieburth L, Voinnet O (2008) Widespread translational inhibition by plant miRNAs and siRNAs. Science 320:1185-1190
-
Goeres DC, Van Norman JM, Zhang W, Fauver NA, Spencer ML, Sieburth LE (2007) Components of the Arabidopsis mRNA Decapping Complex Are Required For Early Seedling Development. Plant Cell 19:1549-1564
-
Van Norman JM, Sieburth LE (2007) Dissecting the Biosynthetic Pathway for the bypass1 Root-Derived Signal. Plant J. 49:619-628
-
Sieburth LE, Muday GK, King EJ, Benton G, Kim S, Metcalf KE, Meyers L, Seamen E (2006) SCARFACE encodes an ARF-GAP that is required for normal auxin efflux and vein patterning in Arabidopsis. Plant Cell 18:1396-1411
-
Van Norman JM, Frederick R, Sieburth L (2004) BYPASS1 negatively regulates a root-derived signal that controls plant architecture. Current Biology 14:1739-1746
-
Deyholos MK, Cavaness GF, Hall B, King E, Punwani J, Van Norman J, Sieburth LE (2003) VARICOSE, a WD-domain protein, is required for leaf blade development. Development 130:6577-6588
-
Turner S, Sieburth LE (December 2002) Vascular Patterning. The Arabidopsis Book, eds. C.R. Somerville and E.M. Meyerowitz, American Society of Plant Biologists, Rockville, MD, doi/10.1199/tab.0073 http://www.aspb.org/publications/arabidopsis/
-
Saulsberry A, Martin PR, O’Brien T, Sieburth LE, Pickett BG (2002) The Induced Sector Arabidopsis Embryonic Fate Map. Development129:3403-3410
-
Deyholos MK, Cordner G, Beebe D, Sieburth LE (2000) The SCARFACE Gene is Required for Cotyledon and Leaf Vein Patterning. Development 127:3205-3213
