Ben Myers
Associate Professor of Oncological Sciences and
Adjunct Associate Professor of Biomedical Engineering and of Biochemistry
Signaling across the Membrane in Development and Cancer: Hedgehog, Wnt, Primary Cilium, Membrane Proteins / Lipids, G protein-coupled receptors

Molecular Biology Program
Biological Chemistry Program
Education
A.B. Harvard University
Ph.D. University of California, San Francisco
Research
In multicellular organisms, cells need to communicate with one another to ensure proper tissue and organ development, and to prevent diseases such as cancer. My lab studies an essential but poorly understood aspect of this process: how signals are transmitted from the outside of the cell across the membrane to the cell interior. We are tackling this problem from an interdisciplinary perspective, drawing on protein and lipid biochemistry, cell biology, physiology, embryology, and a range of related approaches.
A better understanding of cell-cell communication will allow us to understand at a molecular level how tissues and organs develop. It will also help us to design better therapies for birth defects, cancers, and other diseases.
Current research in my lab focuses on:
- Signal transduction in the Hedgehog cascade, which controls the development of nearly all of our tissues and organs and is mutated in several widespread cancers. Surprisingly, we know nearly nothing about how some of the most critical steps in this pathway operate. We are approaching this question by merging in vitro biochemistry, biophysics, and structural biology with in vivo cellular and organismal functional readouts. Our studies have provided foundational insights into how Hedgehog signals are received and transmitted across the membrane. In turn, these insights are helping us develop innovative strategies to control this pathway in cancer and degenerative disorders.
- Signaling pathways in the primary cilium. The primary cilium is a tiny antenna-shaped membrane protrusion that is critical to Hedgehog and a range of other signaling pathways controlling the nervous, cardiovascular, and musculoskeletal systems. We are using novel live cell fluorescent activity reporters to dissect the signaling events that unfold within this tiny organelle.
- How lipids and kinases influence signal transduction. Our work on Hedgehog has led to new principles regarding how lipids can bind to and activate membrane receptors, and how membrane receptors can signal to kinases. We are now using these principles to unravel the signaling mechanisms in a variety of other signaling pathways, including G protein-coupled receptor cascades critical throughout human health and physiology.
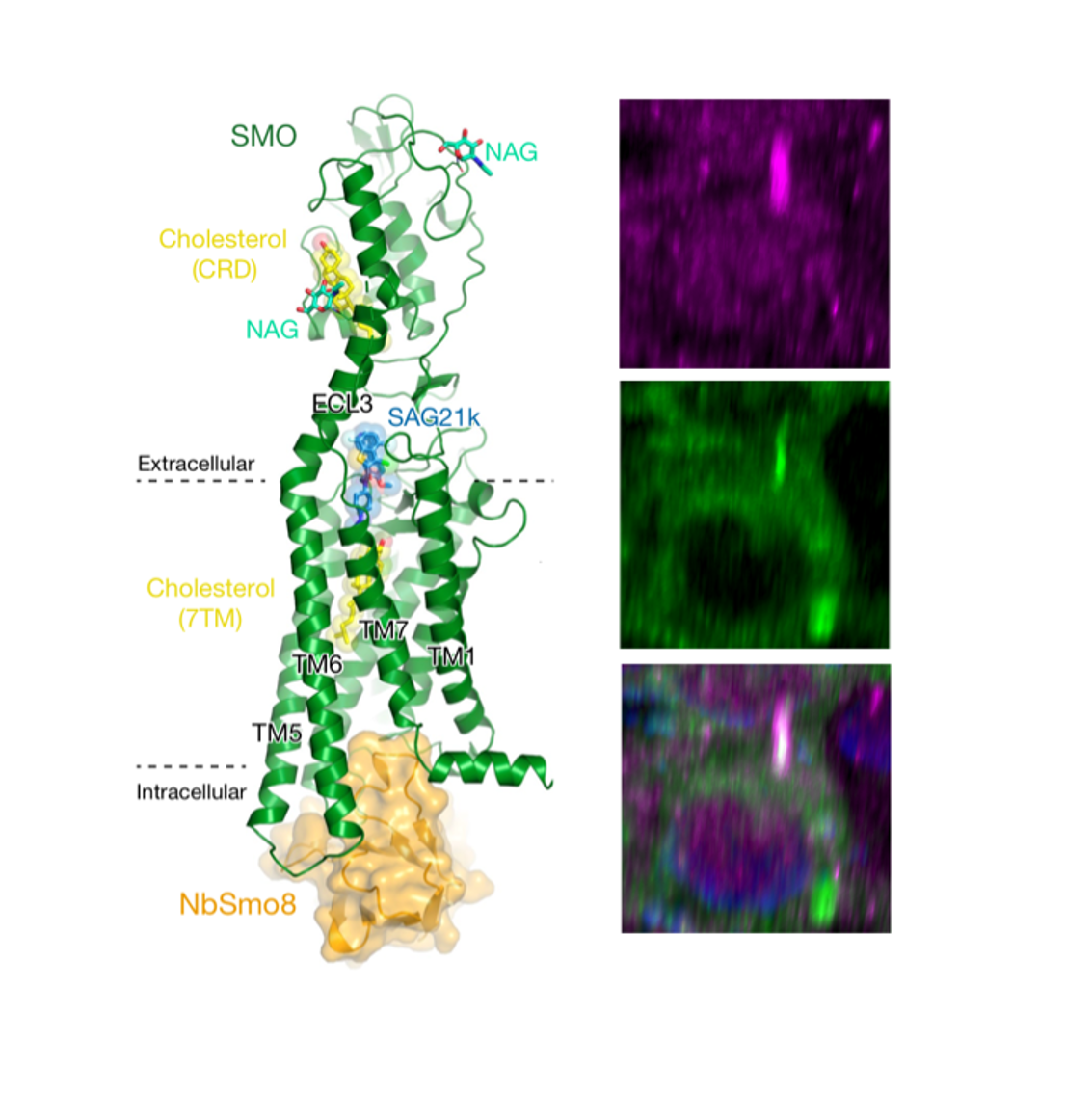
Left: A 2.8 Å crystal structure of SMOOTHENED (green), the main signal transducer in the Hedgehog pathway, bound to cholesterol ligands (yellow) and trapped in an active state by a conformation-specific nanobody (NbSmo8, orange)(Desphande et al, Nature 2019). Right: Hedgehog signaling in the primary cilium. SMOOTHENED (magenta) controls protein kinase A (PKA, green) via an unprecedented mechanism - it directly binds and inactivates PKA by physically blocking the enzyme’s active site (Arveseth et al, PLOS Biol 2021; Happ et al, Nat Struct Mol Biol 2022.). The cilium is marked by SMOOTHENED staining (magenta), and the nucleus is marked by Hoechst staining (blue).
References
- Steiner WP, Iverson N, Venkatakrishnan V, Wu J, Stepniewski TM, Michaelson Z, Bröckel JW, Zhu J-F, Bruystens J, Lee A, Nelson I, Bertinetti D, Arveseth CD, Tan G, Spaltenstein P, Xu J, Hüttenhain R, Kay M, Herberg FW, Selent J, Anand GS, Dunbrack RL*, Taylor SS*, Myers BR*. A structural mechanism for noncanonical GPCR signal transduction in the Hedgehog pathway. bioRxiv [Preprint]. 2024 Nov 1:2024.10.31.621410. doi: 10.1101/2024.10.31.621410. PMID: 39554190; PMCID: PMC11565934. (* = corresponding author)
- Walker MF, Zhang J, Steiner W, Ku P-I, Zhu J-F, Michaelson Z, Yen Y-C, Lee A, Long AB, Casey MJ, Poddar A, Nelson IB, Arveseth CD, Nagel F, Clough R, LaPotin S, Kwan KM, Schulz S, Stewart RA, Tesmer JJG, Caspary T, Subramanian R*, Ge X*, Myers BR*. GRK2 kinases in the primary cilium initiate SMOOTHENED-PKA signaling in the Hedgehog cascade. PLOS Biology 2024 Aug 13; 22(8). doi: 10.1371/journal.pbio.3002685. eCollection 2024 Aug. PMID: 39138140. (* = corresponding author).
- Zhang J, Kaur G, Cai E, Gutierrez OT, Liu X, Baboo S, Diedrich JK, Zhu JF, Myers BR, Yates JR 3rd, Ge X. Proximity based proteomics reveals Git1 as a regulator of Smoothened signaling [Internet]. bioRxiv [Preprint] .2025. Available from: http://dx.doi.org/10.1101/2025.01.06.631593 PMCID: PMC11741329
- Hilgendorf KI, Myers BR, Reiter JF. (2024). Emerging mechanistic understanding of cilia function in cellular signalling. Nature Reviews Molecular and Cell Biology. 1–19.
- Happ JT*, Arveseth CD*, Bruystens J*, Bertinetti D*, Nelson IB*, Olivieri C*, Zhang J, Hedeen DS, Zhu J, Capener JL, Brockel JW, Vu L, King CC, Ruiz-Perez VL, Ge X, Veglia G, Herberg FW, Taylor SS^, Myers BR^. A PKA Inhibitor Motif within Smoothened Controls Hedgehog Signal Transduction. Nature Structural and Molecular Biology 2022 Oct; 29(10):990-999. doi: 10.1038/s41594-022-00838-z. Epub 2022 Oct 6. (* = co-first author; ^ = co-corresponding author). PMCID: PMC9696579.
- Arveseth CD*, Happ J*, Hedeen D, Zhu J, Capener J, Shaw D, Deshpande I, Liang J, Xu J, Stubben S, Nelson IB, Walker M, Krogan N, Grunwal D, Hüttenhain R, Manglik A, Myers BR. Smoothened Transduces Hedgehog Signals via Activity-Dependent Sequestration of PKA Catalytic Subunits. PLoS Biology 19(4): e3001191 (2021) (* = equal contribution)
- Deshpande I, Liang J, Hedeen D, Roberts KJ, Zhang Y, Ha B, Latorraca NR, Faust B, Dror RO, Beachy PA, Myers BR^, Manglik A^. Smoothened Stimulation by Membrane Sterols Drives Hedgehog Pathway Activation. Nature (2019) https://doi.org/10.1038/s41586-019-1355-4 (^ = corresponding author)
- Zhang Y, Bulkley DP, Xin Y, Roberts KJ, Asarnow DE, Sharma A, Myers BR, Cho W, Cheng Y, Beachy PA. Structural Basis for Cholesterol Transport-like Activity of the Hedgehog Receptor Patched. Cell, Nov 15;175(5):1352-1364.e14 (2018)
- Myers BR*^, Neahring L*, Zhang Y, Roberts KR, Beachy PA^. Rapid, direct activity assays for Smoothened reveal Hedgehog pathway regulation by membrane cholesterol and extracellular sodium. Proc Nat Acad Sci10.1073/pnas.1717891115 (2017) (* = first author; ^ = corresponding author). pdf
- Commentary by Blassberg and Briscoe in Dev Cell: pdf
- Sweeney RT*, McClary AC*, Myers BR*, Biscocho J*, et al. Identification of Recurrent SMO and BRAF Mutations in Ameloblastomas. Nat Gen46(7):722-5 (2014). (* = first author)
- Myers BR, Sever N, Chong YC, Kim J, Belani JD, Rychnovsky S, Bazan JF, Beachy PA. Hedgehog Pathway Modulation by Multiple Lipid Binding Sites on the Smoothened Effector of Signal Response. Dev Cell 26(4):346-57 (2013).
- Myers BR, Sigal YM, Julius D. Evolution of Thermal Response Properties in a Cold-Activated TRP Channel. PLoS ONE 4(5): e5741. doi:10.1371/journal.pone.0005741 (2009).
- Prober DA, Zimmerman S, Myers BR, McDermott BM, Caron S, Rihel J, Kim S, Kettleborough RNW, Stemple DL, Solnica-Krezel L, Julius D, Hudspeth AJ, Schier AF. Zebrafish TRPA1 Channels are Required for Behavioral Responses to Mustard Oil but not for Thermosensation or Mechanosensory Hair Cell Function. J Neurosci 28(40):10102-10 (2008).
- Myers BR, Saimi Y, Kung C, Julius D. Multiple Unbiased Prospective Screens Identify TRP Channels and their Conserved Gating Elements. J Gen Physiol. 132(5): 481-6 (2008).
- Myers BR, Bohlen C, Julius D. A Yeast Genetic Screen Reveals a Critical Role for the Pore Helix Domain in TRP Channel Gating. Neuron 58(3):362-73 (2008).
- Myers BR, Julius D. TRP Channel Structural Biology: New Roles for an Old Fold. Neuron 54(6):847-50 (2007).
- Desai BN, Myers BR, Schreiber SL. FKBP12-rapamycin associated protein associates with mitochondria and senses osmotic stress via an intermediate mitochondrial dysfunction. Proc. Natl. Acad. Sci. USA 99(7):4319-24 (2002).
