Matthew Miller
Assistant Professor of Biochemistry
Chromosome Segregation, Cell Division, Microtubules, Cell Biology, Biochemistry, Biophysics

Molecular Biology Program
Biological Chemistry Program
Education
B.A. Carleton College
Ph.D. Massachusetts Institute of Technology
Research
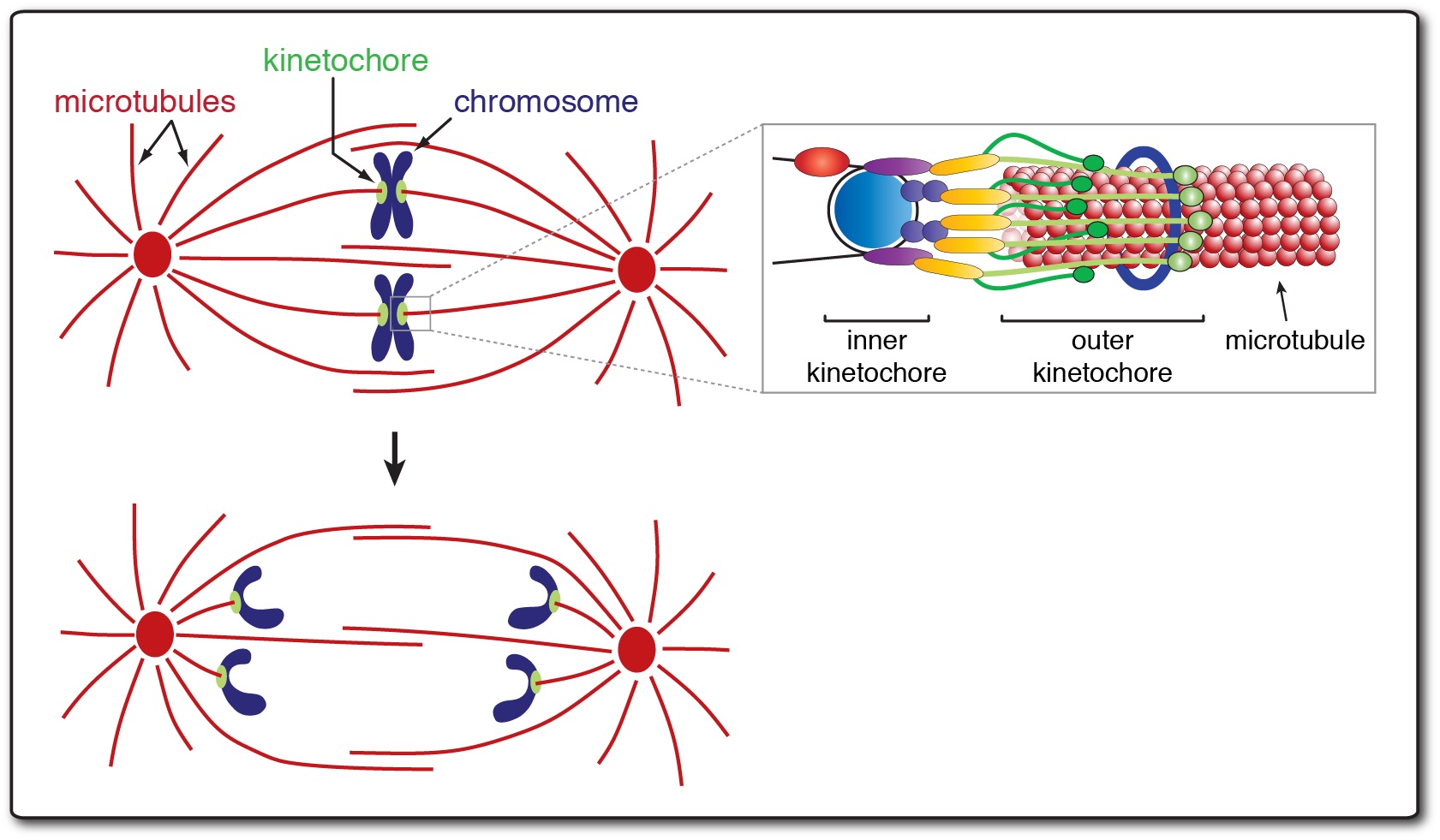
The accurate segregation of chromosomes during cell division is fundamental to cellular and organismal fitness. Errors in this process are the leading cause of miscarriages and congenital birth defects and are the most prevalent genetic alteration in tumor cells. Despite this, we know very little about why chromosome segregation is so defective in these circumstances. Chromosome segregation is mediated by a highly conserved protein complex, the kinetochore, which physically attaches chromosomes to spindle microtubules to pull the chromosomes apart. Kinetochores are incredible protein machines. They move chromosomes by remaining persistently attached to their constantly changing substrate (i.e. dynamically growing and shrinking microtubule tips). They are also signaling hubs, sensing and halting the cell cycle when they’re improperly attached and self-correcting these improper attachments. Although biologists have been fascinated with this process for over a hundred years, we still do not know how the kinetochore achieves these astonishing feats. Our lab takes an interdisciplinary approach, utilizing in vitro reconstitution-based assays combined with yeast genetics and cell biology to understand the macromolecular machines that carry out the process of chromosome segregation.
For more information please see our lab website at https://www.miller.biochem.utah.edu/research
References
- Zahm, J. A., Stewart, M. G., Carrier, J. S., Harrison, S. C., Miller, M.P. Structural Basis of Stu2 Recruitment to Yeast Kinetochores. eLife (2021) 10:e65389
- Gutierrez, A., Kim, J. O., Umbreit, N. T., Asbury, C.L., Davis, T. N., Miller, M.P., Biggins, S. Cdk1 Phosphorylation of the Dam1 Complex Strengthens Kinetochore-Microtubule Attachments. Curr Bio (2020) 30(22):4491-4499.e5
- Herman, J.A., Miller, M.P., Biggins, S. chTOG is a conserved mitotic error correction factor eLife (2020) 9:e61773
- Miller, M. P., Evans, R. K., Zelter, A., Geyer, E. A., Rice, L. M., Davis, T. N., Asbury, C. L., Biggins, S. A Stu2-mediated intrinsic tension-sensing pathway promotes chromosome biorientation in vivo. PLoS Genet (2019) 15(10):e1008423
- Geyer, E. A., Miller, M. P., Brautigam, C. A, Biggins, S., Rice, L. M., Design principles of a microtubule polymerase eLife (2018) Jun 13;7. pii: e34574.
- Miller, M. P., Asbury, C. L. & Biggins, S. A TOG Protein Confers Tension Sensitivity to Kinetochore-Microtubule Attachments. Cell 165(6):1428-1439 (2016).
- Umbreit, N. T., Miller, M. P., et al. Kinetochores require oligomerization of Dam1 complex to maintain microtubule attachments against tension and promote biorientation. Nat. Commun. 5, 4951 (2014).
- Attner, M. A., Miller, M. P., Ee, L.-S., Elkin, S. K. & Amon, A. Polo kinase Cdc5 is a central regulator of meiosis I. Proc. Natl. Acad. Sci. U. S. A. 110(35):14278-83 (2013).
- Miller, M. P., Amon, A. & Unal, E. Meiosis I: when chromosomes undergo extreme makeover. Curr. Opin. Cell Biol. (2013) 25(6):687-96.
- Miller, M. P.*, Unal, E.*, Brar, G. A. & Amon, A. Meiosis I chromosome segregation is established through regulation of microtubule-kinetochore interactions. eLife 1, e00117 (2012). (*) equal contribution
- Miller M.P., Suominen P, Aristidou A, Hause B, Van Hoek P, Asleson Dundon C. Lactic acid-producing yeast cells having nonfunctional l- or d-lactate:ferricytochrome c oxidoreductase cells, United States Patent (2012) 8,137,953.
