Nitin Phadnis
Associate Professor of Biological Sciences
Speciation, Genetic Conflict Segregation Distortion, Chromosome Mechanics

Molecular Biology Program
Education
Research
In his seminal work “On the Origin of Species”, Darwin presented overwhelming evidence for two fundamental phenomena. First, species adapt; and second, new species arise from existing species. A combination of these two processes provides a natural explanation for the origins of the staggering diversity of life. While Darwin explained the first phenomenon – adaptation – through the elegant mechanism of natural selection, he could find no satisfactory explanation to the second phenomenon – speciation – and termed it the “mystery of mysteries”. This “mystery of mysteries” continues to be one of the most important and unsolved problems in biological research over the past 150 years: How do two species evolve from one? The Phadnis Laboratory takes a multi-disciplinary approach combining genetics, genomics and cell biology to address this “mystery of mysteries” and to understand the evolutionary forces that shape the structures of genomes.
1. The Molecular Basis of Speciation
Speciation – the process by which one species splits into two – involves the evolution of reproductive isolating barriers such as the sterility or inviability of hybrids between populations. We would like to understand the molecular processes which, when tinkered by evolution, lead to a breakdown in hybrids and have profound consequences for speciation. Despite decades of intense research, we still know of very few genes that cause the sterility or inviability of hybrids. The extreme difficulty in identifying these genes arises from a methodological paradox. Genetics involves crosses and species, by definition, don’t readily cross.
The Phadnis laboratory specializes in identifying the genes that cause hybrid sterility or inviability in various Drosophila species. In particular, we are interested in identifying the genes that are important for hybrid dysfunction in evolutionarily young species that are in the process of ongoing speciation, such as the Bogota and USA subspecies of Drosophila pseudoobscura. We further use cell biological methods to dissect the molecular pathways that are disrupted in hybrids and apply evolutionary approaches to identify the biological causes of divergence at these genes. Finally, we are interested in developing powerful genomics-based approaches to rapidly hybrid sterility and inviability genes in other model and non-model organisms.
2. Intra-Genomic Evolutionary Conflict
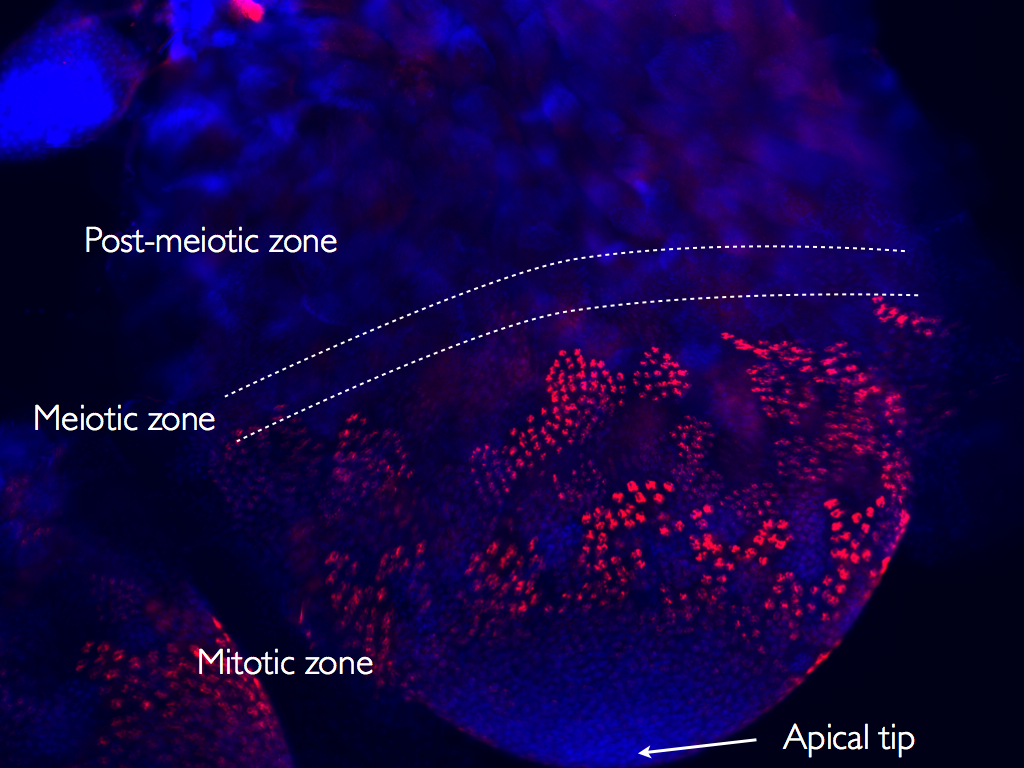
Our genomes, instead of being static entities, are dynamic collectives of genes that are in evolutionary conflict with each other. For example, segregation distorters are selfish genetic elements that subvert the mechanisms of meiosis to over-represent themselves in gametes, thus gaining a massive evolutionary advantage. Segregation distorters enjoy their evolutionary advantage even when they impose a fitness cost on the individual carrying the distorter alleles, and thus provide selection pressure for the evolution of suppressors of segregation distortion. Continuing molecular arms races between segregation distorter genes and their suppressors can lead to the rapid evolution of meiosis genes, and in turn cause them to become functionally incompatible between species.
These enigmatic selfish elements, which work by violating Mendel’s Laws, are ubiquitous in nature and are a potent evolutionary force in influencing the structures of genomes and in the evolution of species. We are interested in identifying the genes and molecular mechanisms deployed by these elements to subvert meiosis. These studies shed light on the evolutionary genetics of such conflicts and also on the inner workings of the meiotic machinery. Current projects include studying the molecular basis of segregation distortion in D. pseudoobscura Bogota-USA hybrids and in the classic SD system in D. melanogaster. Most interestingly, we are engaged in uncovering evidence of genetic conflict involving segregation distorters as a driving force in the evolution of hybrid sterility during the earliest stages of speciation.
3. The Causes and Consequences of Rapid Evolution
Advances in whole genome sequencing and population genetic methods have revealed several surprising classes of genes that evolve rapidly under positive selection. While detecting selection at genomic locations has become easier, a detailed understanding of the evolutionary causes and the functional consequences of the rapid divergence of genes is often missing. These interesting classes of genes include those that are involved in meiosis, nuclear transport, dosage compensation, RNA interference, etc.
We are interested in understanding the causes of rapid evolution at these genes, and the molecular consequences of their functional divergence. Our laboratory uses the formidable collection of mutants available in D. melanogaster and well developed cytological techniquesto ask whether the molecular functions of such genes can be replaced by transgenically-introduced non-adapted alleles from its sister species D. simulans. The molecular analyses of such non-complementing genes and the phenotypes revealed during inter-species swaps provide an exciting new window into understanding the causes of positive selection at these genes and into uncovering the divergence of molecular functions of genes that evolve rapidly between species.
Taken together, these research projects are helping unravel Darwin's "mystery of mysteries" by informing the molecular and functional basis of speciation and by guiding the investigation of fundamental evolutionary processes that shape the complexity of genomes, cells, and species.
References
-
Cooper, J.C. & Phadnis, N. (2017) Parallel evolution of sperm hyper-activation Ca2+ channels. Genome Biology and Evolution doi:10.1093/gbe/evx131
-
Cooper, J.C. & Phadnis, N. (2016) A genomic approach to identify hybrid incompatibility genes. Fly, 26:1-7
-
Phadnis, N., Baker, E.P., Cooper, J.C., Frizzell, K., Hseih, E., de la Cruz, A.F., Shendure, J, Kitzman, J., and Malik, H.S. (2015) An essential cell cycle regulation gene causes hybrid inviability in Drosophila.Science, 350: 1552-1555.
-
Phadnis, N. and Malik H.S. (2013) The Molecular and Evolutionary Basis of Hybrid Sterility: From Odysseus to Overdrive. Chapter 9 in Speciation: Natural Processes, Genetics and Biodiversity. Ed. Michalak, P., Nova Science Publishers, Inc.: 207-226.
-
Phadnis, N., Hsieh, E. and Malik, H.S. (2011) Birth, death and replacement of karyopherins in Drosophila. Molecular Biology and Evolution 29(5): 1429-40.
-
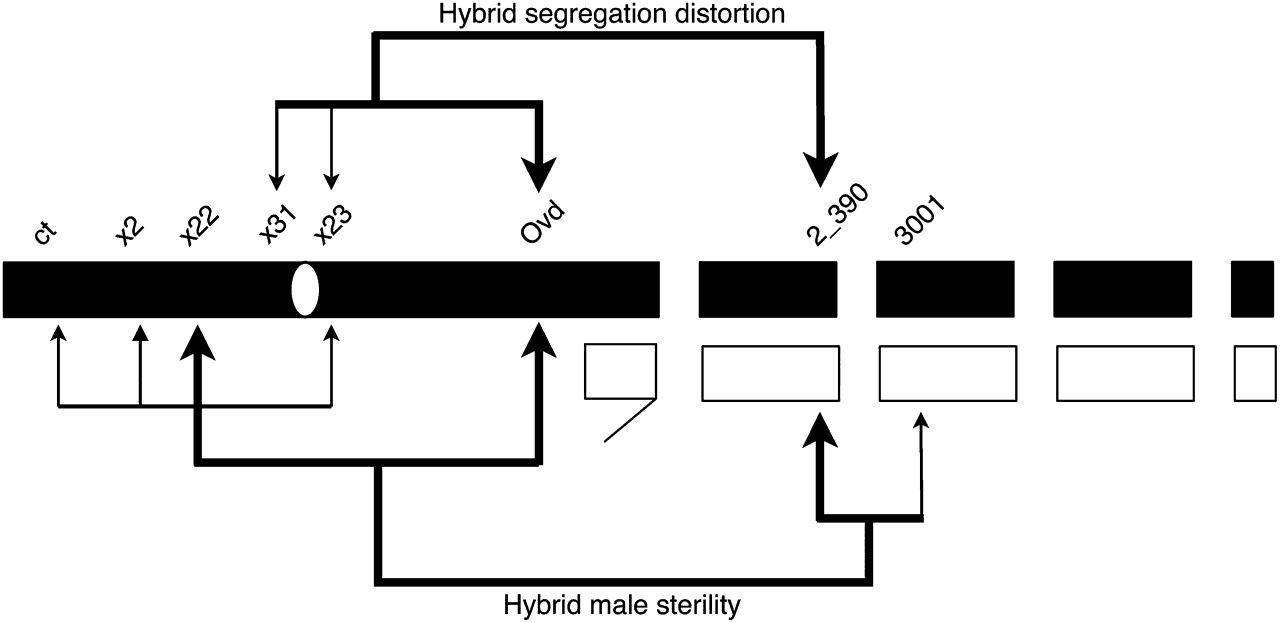
Phadnis, N. (2011) Genetic architecture of male sterility and segregation distortion in Drosophila pseudoobscura Bogota-USA hybrids. Genetics 189(3):1001-1009.
-
Oliver, P.L., Goodstadt L, Bayes JJ, Birtle, Z., Roach KC, Phadnis, N, Beatson SA, Lunter, G, Malik, H.S., and Ponting, C.P. (2009) Accelerated evolution of the Prdm9 speciation gene across diverse metazoan taxa. PLoS Genetics 5(12):e1000753. (Media attention: Faculty of 1000: http://f1000.com/1304956)
-
Phadnis, N. and Orr, H.A. (2009) A single gene causes both male sterility and segregation distortion in Drosophila hybrids. Science 323(5912): 376-378. (Media attention: Science: Origin of species in Overdrive. Faculty of 1000: http://f1000.com/1160563; Nature News: The genes that drive speciation.; Nature Reviews Genetics: A bumper crop for speciation genetics.; Science News: Evolution’s wedges: Finding the genes that drive one species into two; Digital Journal: The first speciation gene in mammals has just been discovered.)
-
Orr, H.A., Masly, J.P and Phadnis, N. (2007) Speciation in Drosophila: from phenotypes to molecules. Journal of Heredity 98(2): 103-110.
-
Phadnis, N. and Fry, J.D. (2005) Widespread correlations between dominance and homozygous effects of mutations: Implications for theories of dominance. Genetics 171(1): 385-392.
-
Fry, J.D., Bahnck, C.M., Mikucki, M., Phadnis, N., and Slattery, W.C. (2004) Dietary Ethanol Mediates Selection on Aldehyde Dehydrogenase Activity in Drosophila melanogaster. Integrative and Comparative Biology 44(4): 275-283.