Michelle Mendoza
Associate Professor of Oncological Sciences and Adjunct Associate Professor of Biomedical Engineering
Cell Migration, Invasion, RAS, ERK, Lung Cancer, Signaling, Tumor Mechanics

Molecular Biology Program
Education
B.S. Pennsylvania State University
Ph.D. University of California, San Diego
Research
The Mendoza lab mission is to help patients with lung cancer and other solid tumors through the discovery of fundamental mechanisms of cancer progression. We apply physical science perspectives to probe the spatial and temporal order of biochemical and mechanical signaling in tumor cells and their microenvironment.
We are interested in the signaling of cell migration and cancer invasion. Questions addressed in the lab include:
- How do signaling pathways synergistically control actin and adhesion dynamics for forward motion?
- How does oncogene activation of the ERK kinase enable invasive plasticity?
- How does lung cancer ERK signaling interact with mechanical changes in the tumor microenvironment to drive progression?
We approach these and other biological problems with a combination of experimental and computational techniques, including biochemistry, quantitative imaging of cell lines and tumor tissue from patients and mouse models, and computational modeling.
Cell movement involves cycles of leading edge protrusion and adhesion to the extracellular substrate, followed by de-adhesion and contraction of the cell body and rear. Actin assembles against the plasma membrane, generating pushing force that induces protrusions. Nascent adhesions assemble at the cell edge for traction force and undergo regulated disassembly versus maturation to balance forward motion and mechanical tension.
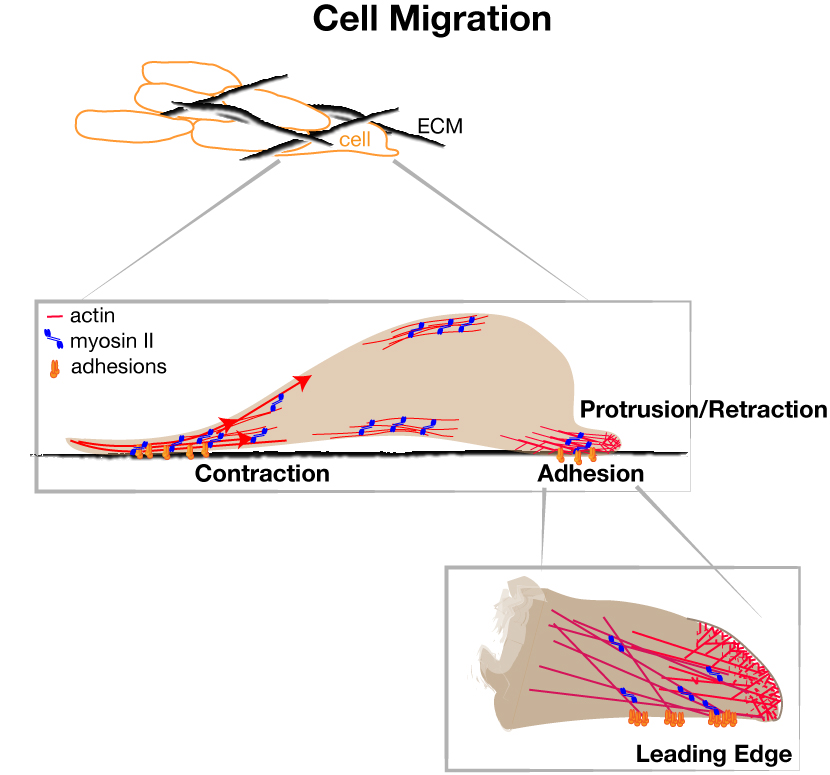
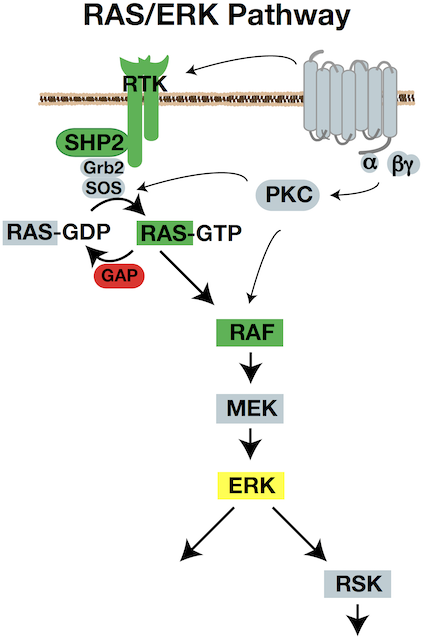
Signaling pathways impinge on the intrinsic cytoskeleton cycles to control the proficiency of movement. The RAS/RAF/MEK/ERK signaling pathway is a key regulator of cell growth, proliferation, survival, differentiation, and also motility. It is also one of the most commonly activated pathways in solid tumors. We aim to understand ERK’s control of cell movement in normal cells, and the consequences of ERK misregulation in cancer dissemination.
Integrated Forces for Cell Migration
We have established that ERK acts on the actin assembly machinery to directly promote rapid and sustained edge protrusion during cell movement. ERK phosphorylates the WAVE Regulatory Complex, which promotes WAVE’s interaction with, recruitment, and activation of the ARP2/3 actin nucleator at the cell edge. This increases actin assembly rates, which generates the pushing force needed to move the membrane forward. We have also found that ERK controls myosin II activity through activation of p90RSK, which phosphorylates the myosin phosphatase regulatory subunit. MB students Andrew Shepherd and Akib Khan are investigating how ERK also controls traction force and membrane tension. Our collaborative work with the Bidone Lab in Biomedical Engineering is testing how coordinate regulation of these molecular forces generates edge protrusion.
ERK and Cancer Invasion
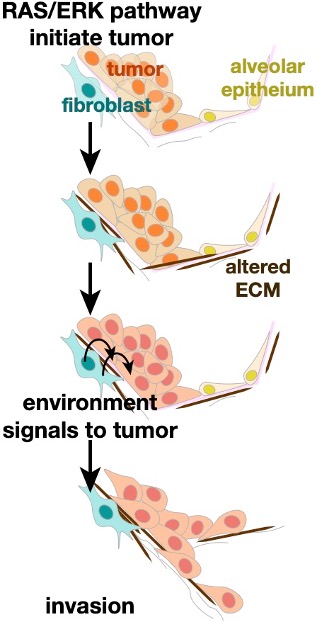
When cancers spread throughout the body, tumor cells navigate altered tumor microenvironments and foreign structures. Biomedical engineering student Becca Zitnay is probing how strain, stiffness, and chemical signals in the tumor microenvironment promote invasion. We are characterizing these signals by probing clinical and animal samples of lung cancer. Through a collaborative project with the Weiss lab in Biomedical Engineering, we are identifying how the altered mechanical forces promote tumor invasion.
Lung Cancer
Oncogenic ERK signaling is essential for lung adenocarcinoma initiation, maintenance, progression into invasive, metastatic disease. While common RAS and RAF mutations activate ERK, additional events are needed to bypass feedback loops and allow hyperactivation. Shiela Samson and Becca Zitnay are working to identify these additional ERK-activating events in lung cancer and how ERK signals for lung cancer invasion in vivo. We apply live-cell imaging of tumor invasion and biosensors in native tissue slices.
References
- Shiela C. Samson, Akib M. Khan, Michelle C. Mendoza. ERK signaling for cell migration and invasion.Front Mol Biosci. 9:998475. (2022).
- Rebecca G.Zitnay, Michael Herron, Keith R.Carney, Scott Potter, Lyska L. Emerson, Jeffrey A. Weiss, Michelle C. Mendoza. Mechanics of lung cancer: A finite element model shows strain amplification during early tumorigenesis. PLoS Comput Biol. 18(10):e1010153. (2022).
- Keith R. Carney, Akib M. Khan, Shiela C. Samson, Nikhil Mittal, Sangyoon J. Han, Michelle C. Mendoza*, and Tamara C. Bidone*. Nascent adhesions differentially regulate lamellipodium velocity and persistence.BioRxiv (2021).
- Kelley Ingram, Shiela C. Samson, Rediet Zewdu, Rebecca G. Zitnay, Eric L. Snyder, and Michelle C. Mendoza. NKX2-1 controls lung cancer progression by inducing DUSP6 to dampen ERK activity. Oncogene. 41(2):293-300 (2022). PMC8738158
- Rediet Zewdu, Elnaz M. Mehrabad, Kelley Ingram, Pengshu Fang, Katherine L. Gillis, Soledad A. Camolotto, Grace G. Orstad, Alex Jones, Michelle C. Mendoza, Benjamin T. Spike, and Eric L. Snyder. An NKX2-1/ERK/WNT feedback loop modulates gastric identity and response to targeted therapy in lung adenocarcinoma. ELife. 10:e66788 (2021). PMC8102067.
- David A. Kircher, Kirby A. Trombetti, Mark R. Silvis, Gennie L. Parkman, Grant M. Fischer, Stephanie M. Angel, Christopher M. Stehn, Sean C. Strain, Allie H. Grossmann, Keith L. Duffy, Kenneth M. Boucher, Martin McMahon, Michael A. Davies, Michelle C. Mendoza, Matthew W. VanBrocklin, and Sheri L. Holemn. AKT1E17K activates focal adhesion kinase and promotes melanoma brain metastasis. Molecular Cancer Research 17(9):1787-1800 (2019).
- Shiela C. Samson, Andrew Elliott, Brian D. Mueller, Yung Kim, Keith R. Carney, Jared P. Bergman, John Blenis, and Michelle C. Mendoza. p90 Ribosomal S6 Kinase (RSK) Phosphorylates Myosin Phosphatase and thereby Controls Edge Dynamics during Cell Migration. J Biol Chem. 294(28):10846-10862. (2019).
- Michelle C. Mendoza*, Marco Vilela*, Jesus E. Juarez, John Blenis, and Gaudenz Danuser. ERK Reinforces Actin Polymerization to Power Persistent Edge Protrusion during Motility.Science Signaling 8(377):ra47 (2015). PMC4830495
- Emrah Er, Michelle C. Mendoza, Ashley M. Mackey, Lucia E. Rameh, and John Blenis. AKT Facilitates EGFR Trafficking and Degradation by Phosphorylating and Activating PIKfyve. Science Signaling 6(279):ra34 (2013). PMC4041878.
- Michelle C. Mendoza. Phosphoregulation of the WAVE Regulatory Complex and Signal Integration.Trends in Biomedical Sciences 24(4):272-9 (2013). PMC3637877.
- Wenjuan Zhang, Michelle C. Mendoza, Xiaolei Pei, Didem Ilter, Sarah J. Mahoney, Yingmei Zhang, Dalong Ma, John Blenis, and Ying Wang. Down-regulation of CMTM8 Induces Epithelial-to-Mesenchymal-like Changes via c-MET/Extracellular Signal-Regulated Kinase (ERK) signaling. J Biol Chem 287:11850-8 (2012). PMC3320933.
- Michelle C. Mendoza, Sebastien Besson, and Gaudenz Danuser. Quantitative Fluorescent Speckle Microscopy (QFSM) to Measure Actin Dynamics. Current Protocols in Cytometry 62:2.18.1-2.18.25 (2012). PMC3688286.
- Michelle C. Mendoza, E. Emrah Er*, Wenjuan Zhang*, Bryan A. Baliff, Hunter L. Elliott, Gaudenz Danuser, and John Blenis. ERK-MAPK Drives Lamellipodia Protrusion by Activating the WAVE2 Regulatory Complex. Molecular Cell 41:661-71 (2011). PMC3078620.
- Michelle C. Mendoza, E. Emrah Er, and John Blenis. The Ras-ERK and PI3K-mTOR Pathways: Cross-talk and Compensation. Trends in Biomedical Sciences 36:320-8 (2011). PMC3112285.
