Clement Chow
Associate Professor of Human Genetics
Genetic Variation, ER Stress, Disease Modifiers

Molecular Biology Program
Education
B.A. Cornell University
Ph.D. University of Michigan
Research
Our lab is focused on understanding the role of genetic variation on disease outcomes. We employ quantitative and functional tools, in a variety of model organisms, to study how genetic variation impacts basic cellular traits important to human health. Our work in model organisms will help to model and inform studies of genetic variation in the human population. We hope to identify variation in the human population that can lead to more precise, personalized therapies.
Precision Medicine and Rare Disease
Precision medicine seeks to treat patients as individuals. We are beginning to recognize that each patient is unique and disease outcomes can vary greatly. Most Mendelian and complex diseases are impacted by a variety of factors, including genetic background. In order to realize the goals of Precision Medicine, we need to understand how all these factors interact to produce individual outcomes. My lab is focused on understanding how genetic background can contribute to disease outcomes and how we might take advantage of these differences to develop individualized therapies.
Rare diseases pose a unique opportunity to develop precision medicine approaches. Each rare disease only affects a small number of people, but collectively, the 7,000 documented rare diseases affect over 30 million Americans. Recent technological advances in diagnosis have brought rare Mendelian diseases back into the spotlight. Whole genome and exome sequencing have increased the rate of genetic diagnosis. Because approaches to Precision Medicine are simplified when the causative mutation is known, we can focus on understanding the factors that drive variability in rare disease. Principles and approaches developed for rare disease should be translatable to common diseases.
Natural Genetic Modifiers: We are particularly interested in understanding how individual genetic background drives differences in disease penetrance and expressivity. We are undertaking a unique approach by combing genetic variation with classical Drosophila genetics to identify natural genetic modifiers. We are testing the effect of Drosophila disease models on ~200 different genetic backgrounds of the Drosophila Genetic Reference Panel (DGRP). The DGRP is comprised of ~200 inbred strains, such that each strain represents a single wild-derived genome. Thus, the 200 strains capture genetic variation that is present in a natural, wild population. Importantly, we have the whole genome sequence of each strain, allowing phenotype-genotype studies and association analysis to identify modifier genes. Candidate modifiers are then interrogated with appropriate functional studies. We also collaborate with clinicians to screen patient samples for co-segregating modifier alleles that we discover. This approach has the potential to uncover novel modifiers, even in well studied disease models. Some of the diseases that we are applying this approach include retinitis pigmentosa and NGLY1 deficiency.
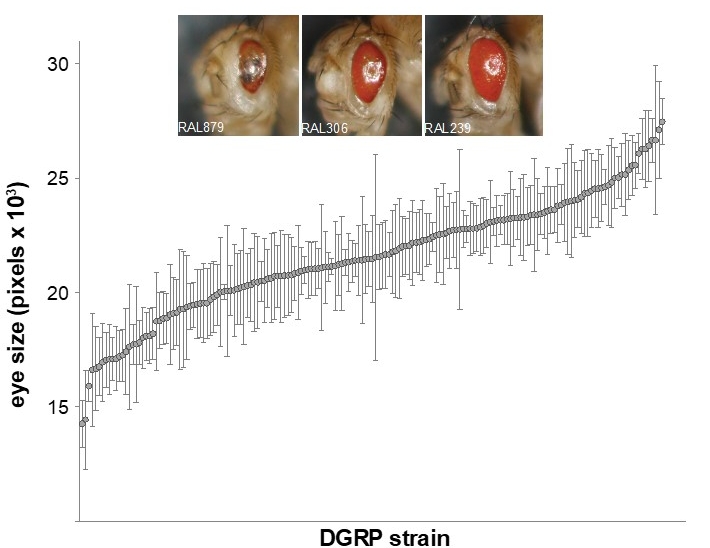
Variation in severity of retinal degeneration in a model of retinitis pigmentosa. Chow et al., 2016
Patient-Specific Drug Screens: Precision medicine often means patients need individualized therapies. To achieve this, we need to build patient-specific models and perform patient-specific small molecule screens. With rare diseases, most of which are severe childhood diseases, this needs to occur very quickly. Drosophila is an ideal tool to achieve this quick transition from the clinic to the bench and back to the clinic for the same patient. We are using either CRISPR or transgenic tools to build patient-specific Drosophila models, in a matter of weeks. These models are then used to perform a repurposing screen with libraries of FDA-approved drugs (i.e. Prestwick Chemical Library), which are key to identifying molecules that can be moved back to the clinic.
Allelic Series: Because there are so few patients with any particular form of rare disease, it can be hard to build genotype-phenotype relationships from clinical reports alone. In some diseases there are “common” disease causing alleles, but in other diseases there can be as many disease causing alleles as there are patients. For a number of these rare disease, we are building patient-specific models. Once we understand the genotype-phenotype relationship, each model can be subjected to a patient specific modifier and small molecule screen.
Genetic Architecture of the ER Stress Response
Genetic variation is a powerful, unbiased tool to uncover novel aspects of gene networks. We are using natural variation to understand how endoplasmic reticulum (ER) stress varies among individuals in a population and what this means for disease variability. To do this, we are using the mouse Collaborative Cross and Diversity Outbred population to uncover the genetic architecture underlying this important cellular stress response. We are also utilizing Drosophila, mouse, and cell culture studies to uncover the function of genes that contribute to variability in the ER stress response.
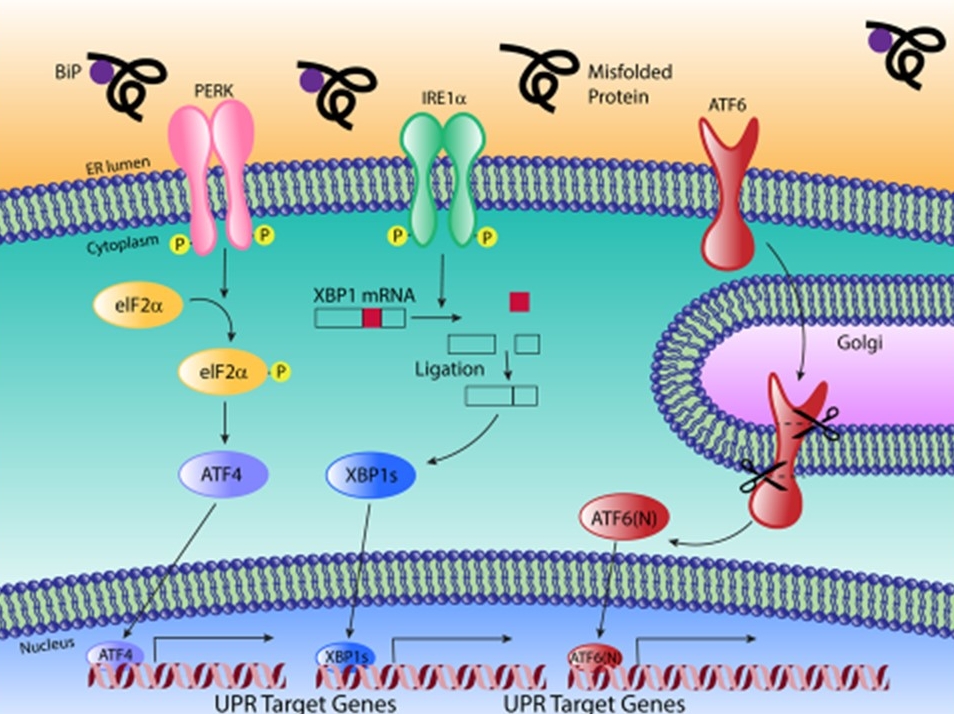
The UPR pathway. Image by Nikki Russell
Evolution
When studying variation, it is impossible to ignore evolution. We are taking an evolutionary approach to understand how different disease genes might act to modify each other. We have projects exploring how different disease genes might coevolve with each other. We are also exploring how evolution has repurposed retrotransposed stress response genes for new functions.
Because natural variation is a powerful tool, we are always open to new ideas.
References
- Talsness DM*, Owings KG*, Coelho E, Mercenne G, Pleinis JM, Zuberi AR, Partha R, Hope KA, Clark NL, Lutz CM, Rodan AR, Chow CY. A Drosophila screen identifies NKCC1 as a modifier of NGLY1 deficiency, eLife. 9: e57831: (2020). *co-first authors
- Palu RAS, Dalton HM, Chow CY. Decoupling of Apoptosis from Activation of the ER Stress Response by the Drosophila Metallopeptidase superdeath. Genetics. 214(4): (2020).
- Ahlers RHL, Trammell CE, Carrell GF, Mackinnon S, Torrevillas BK, Chow CY, Luckhart S, Goodman AG. Insulin potentiates JAK/STAT signaling to broadly inhibit flavivirus replication in insect vectors. Cell Reports. 29(7): (2019).
- Palu RAS, Ong E, Stevens K, Chung S, Owings KG, Goodman AG, Chow CY. Natural genetic variation screen in Drosophila identifies Wnt signaling, mitochondrial metabolism, and redox homeostasis genes as modifiers of apoptosis. G3. 9(12): (2019).
- Palu R, Chow CY. Baldspot/ELOVL6 is a conserved modifier of disease and the ER stress response. PLOS Genet. 14(8): (2018).
- Lavoy S, Chittoor-Vinod VG, Chow CY, Martin I. Genetic Modifiers of Neurodegeneration in a Drosophila Model of Parkinson's Disease. Genetics. 209(4): (2018).
- Owings KG, Lowry JB, Bi Y, Might M, Chow CY. Transcriptome and functional analysis in a Drosophila model of NGLY1 deficiency provides insight into therapeutic approaches. Hum. Mol. Gen. 27(6): (2018).
- Chow CY, Reiter LT. Etiology of Human Genetic Disease on the Fly. Trends Genet. 17: (2017).
- Chow CY. Bringing genetic background into focus. Nature Reviews Genetics. 17(2): (2016).
- Chow CY, Kelsey K, Wolfner MF, Clark AG. Candidate genetic modifiers of retinitis pigmentosa identified by exploiting natural variation in Drosophila. Hum. Mol. Gen. 25(4): (2016).
- Chow CY, Avila F, Wolfner MF, Clark AG. Induction of excessive endoplasmic reticulum stress in the Drosophila male accessory gland results in infertility. PLOS ONE. 10(3): (2015).
- Chow CY, Wang X, Riccardi D, Wolfner MF, Clark AG. The genetic architecture of the genome-wide transcriptional response to ER stress in the mouse. PLOS Genet. 11(2): (2015).
- Chow CY, Wolfner MF, Clark AG. Using natural variation in Drosophila to discover previously unknown endoplasmic reticulum stress genes. PNAS. 110(22): 9013-9018 (2013).
- Chow CY, Wolfner MF, Clark AG. A large neurological component to genetic differences underlying biased sperm use in Drosophila. Genetics. 193(1):177-8 (2013).
- Lenk GM, Ferguson CJ, Chow CY, Jin N, Jones JM, Grant AE, Zolov SN, Winters JJ, Giger RJ, Dowling JJ, Weisman LS, Meisler MH. Pathogenic mechanism of the FIG4 mutation responsible for Charcot-Marie-Tooth disease CMT4J. PLoS Genet. Jun 7(6) (2011).
- Im KM, Kirchhoff T, Wang X, Green T, Chow CY, et al. Haplotype Structure in Ashkenazi Jewish BRCA1 and BRCA2 Mutation Carriers. Hum Genet. 130(5):685-99 (2011).
- Chow CY, Wolfner MF, Clark AG. The Genetic Basis for Male x Female Interactions Underlying Variation in Reproductive Phenotypes of Drosophila. Genetics. 186(4):1355-65 (2010).
- Sirot LK, LaFlamme BA, Stitnik JL, Rubinstein CD, Avila FW, Chow CY, Wolfner, MF. Molecular social interactions: Drosophila melanogaster seminal fluid proteins as a case study. Adv. Genet. 68: 23-56 (2009).
- Chow CY, Landers JE, Bergren SK, Sapp PC, Grant AE, Jones JM, Everett L, Lenk GM, McKenna-Yasek DM, Weisman LS, Figlewicz D, Brown RH, Meisler MH. Deleterious Variants of FIG4, a Phosphoinositide Phosphatase, in Patients with ALS. Am J Hum Genet. 84(1):85-8 (2009).
- Jin N*, Chow CY*, Liu L, Zolov SN, Bronson R, Davisson M, Petersen JL, Zhang Y, Park S, Duex JE, Goldowitz
D, Meisler MH and Weisman LS. VAC14 nucleates a protein complex that is essential
for the regulation of PI(3,5)P2 levels in yeast and mouse. EMBO J. (27), 3221-34 (2008).
*co-first authors - Zhang X, Chow CY, Sahenk Z, Shy M, Meisler M, Li J. Mutation of FIG4 causes a rapidly progressive, asymmetric neuronal degeneration. Brain 131, 1990-2001 (2008).
- McKinney B, Chow CY, Meisler M, Murphy G. Exaggerated emotional behavior in mice heterozygous null for the sodium channel Scn8a (Nav1.6). Genes, brain, and behavior 7 (6), 629-38 (2008).
- Zhang Y, Zolov S, Chow CY, Slutsky S, Richardson S, Piper R, Yang B, Nau J, Westrick R, Morrison S, Meisler M, Weisman L. Loss of Vac14, a regulator of the signaling lipid phosphatidylinositol 3,5-bisphosphate, results in neurodegeneration in mice. PNAS 104 (44), 17518-23 (2007).
- Chow CY, Zhang Y, Dowling J, Jin N, Adamska M, Shiga K, Szigeti K, Shy M, Li J, Zhang X, Lupski J, Weisman L, Meisler M. Mutation of FIG4 causes neurodegeneration in the pale tremor mouse and patients with CMT4J. Nature 448 (7149), 68-72 (2007).
