H. Joseph Yost
Professor of Neurobiology and Adjunct Professor of Pediatrics
Director, Developmental Biology Training Grant
Vice Chairman for Basic Science Research, Department of Pediatrics
Genes and Developmental Mechanisms

Molecular Biology Program
Education
B.S. Creighton University
Ph.D. University of Chicago
Research
All animals start life as a single cell, the fertilized egg, which divides into hundreds of different cell types. Our long-term research goal is to understand the genes, molecules and developmental mechanisms that regulate the assignment of different cell identities in functionally appropriate positions in the developing vertebrate embryo, and to utilize this knowledge for the advancement of human medicine. This top-down approach utilizes embryonic phenotypes to uncover novel genetic pathways and cellular mechanisms ranging from extracellular signaling to genome-wide regulation of transcriptional networks.
Left-Right Axis Formation and Cilia
Our lab is a founder and leader in the field of vertebrate left-right development. Our research utilizes zebrafish, Xenopus and mice to discover the genes, molecular and cellular mechanisms that control asymmetric development of the brain, heart and gut. We have discovered novel mechanisms by which major cell-cell signaling pathways (FGF, TGFbeta and Wnt) control cilia function, cell migration and downstream pathways.
Cardiac Development
We are using genome-wide approaches to elucidate the Gene Regulatory Networks that control heart development in zebrafish, with the goal of understanding human complex congenital cardiac defects that are the leading cause of death in children in the first year. As one of four NHLBI-funded Cardiovascular Development Centers in the country, we have built a team developmental biologists, cardiac physiologists, and experts in chromatin structure, genome-wide gene network profiling, bioengineering and bioinformatics. We have created approaches that exclusively mark either polysomes or whole nuclei in a lineage-specific manor, allowing us to perform genome-wide expression profiling and analysis of the epigenome (RNA-Seq, ChIP-Seq, DNA bisulfite sequencing, etc) from rare cell lineages in the context of a whole animal at any stage of development. In parallel with our zebrafish cardiovascular projects, we have a collaborative project developing human amniocytes, which have nascent stem cell characteristics, into cardiomyocyte lineages with the long-term goal of clinical repair of human congenital heart defects.
Genomics in Zebrafish
Driven by our interest in Gene Regulatory Networks in development and disease, and zebrafish’s reputation for a challenging genome, we have created several molecular and bioinformatics approaches to discovering the molecular lesions of new mutations and rapidly identifying extant mutations. This will allow us to more fully manipulate the zebrafish genome in order to generate models systems of human genetic disorder, including specific cancers and complex heart disease.
The Glycocode Hypothesis
Heparan Sulfate Proteoglycans (HSPGs) are cell surface proteins with long chains of repeating sugar units (glycosaminoglycans; GAGs). Our discovery that a family of HSPGs control cell signaling pathways, cell migration and fibrillogenesis led to our focus on large gene families that regulate proteoglycan biochemistry and function. Our results suggest the existence of a “Glycocode” in which specific sulfation patterns on cell surface GAGs control specific cell-cell signaling decisions, including the modulation of Wnt, BMP and FGF signaling pathways. The Glycocode could provide enormous molecular diversity that dwarfs the informational content of the genome. Discovering out how this Glycocode is regulated and how it is utilized in biology will be one of the major challenges in the “postgenomic era” and will provide important therapeutic targets for a wide range of human diseases.
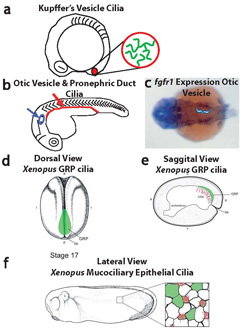
Neugebauer et al, Nature 2009
References
Left-Right, Cilia and Cardiac Development
- Bisgrove BW, Makova S, Yost HJ, Brueckner M (2012) RFX2 is essential in the ciliated organ of asymmetry and an RFX2 transgene identifies a population of ciliated cells sufficient for fluid flow. Developmental Biology, 363:166-78
- Wang G, Cadwallader AB, Jang DS, Tsang M, Yost HJ, Amack JD (2011) The Rho Kinase Rock2b establishes anterior-posterior asymmetry of the ciliated Kupffer's Vesicle in zebrafish. Development, 138:45-54
- Neugebauer JM, Amack JD, Peterson AG, Bisgrove BW, Yost HJ (2009) FGF signaling during embryo development regulates cilia length in diverse epithelia. Nature, 458: 651-4
- Wythe JD, Jurynec MJ, Urness LD, Jones CA, Sabeh MK, Werdich AA, Sato M, Yost HJ, Grunwald DJ, Macrae CA, Li DY (2011) Hadp1, a newly identified pleckstrin homology domain protein, is required for cardiac contractility in zebrafish. Disease Models and Mechanisms, 4:607-21
- Yost HJ (2009) Coordinating the development of bilateral symmetry and left-right asymmetry. Seminars in Cell Dev Biol, 20: 455
- Wang X, Yost HJ (2008) Initiation and propagation of posterior to anterior (PA) waves in zebrafish left-right development. Dev Dyn, 237:3640-7
- Takeuchi JK, Lickert H, Bisgrove BW, Sun X, Yamamoto M, Chawengsaksophak K, Hamada H, Yost HJ, Rossant J, Bruneau BG (2007) Baf60c is a nuclear Notch signaling component required for the establishment of left-right asymmetry. Proc Natl Acad Sci, 104: 846-51
- Amack JD, Wang X, Yost HJ (2007) Two T-box genes play independent and cooperative roles to regulate morphogenesis of ciliated Kupffer's vesicle in zebrafish. Developmental Biology, 310: 196-210
- Sato M, Tsai HJ, Yost HJ (2007) Semaphorin3D regulates invasion of cardiac neural crest cells into the primary heart field. Developmental Biology, 298: 12-21
Zebrafish Genomics and Modeling Human Diseases
- Lezin G, Kosaka Y, Yost HJ, Kuehn MR, Brunelli L (2011) A one-step miniprep for the isolation of plasmid DNA and lambda phage particles. PLoS One, 6: 23457
- Parant JM, George SA, Holden JA, Yost HJ (2010) Genetic Modeling of Li-Fraumeni Syndrome in zebrafish. Disease Models and Mechanisms, 3:45-56
- Parant JM, George SA, Pryor R, Wittwer CT, Yost HJ (2009) A rapid and efficient method of genotyping zebrafish mutants. Dev Dyn, 238:3168-74
- Tsai IC, Woolf M, Neklason DW, Branford WW, Yost HJ, Burt RW, Virshup DM (2007) Disease-associated casein kinase I delta mutation may promote adenomatous polyps formation via a Wnt/beta-catenin independent mechanism. Int J Cancer, 120: 1005-12
- Tsai IC, Amack JD, Gao ZH, Band V, Yost HJ, Virshup DM (2007) A Wnt-CKIvarepsilon-Rap1 pathway regulates gastrulation by modulating SIPA1L1, a Rap GTPase activating protein. Developmental Cell, 12:335-47
- Luo W, Peterson A, Garcia BA, Coombs G, Kofahl B, Heinrich R, Shabanowitz J, Hunt DF, Yost HJ, Virshup DM (2007) Protein phosphatase 1 regulates assembly and function of the beta-catenin degradation complex. EMBO J, 26:1511-21
- Kwan KM, Fujimoto E, Grabher C, Mangum BD, Hardy ME, Campbell DS, Parant JM, Yost HJ, Kanki JP, Chien CB (2007) The Tol2kit: a multisite gateway-based construction kit for Tol2 transposon transgenesis constructs. Dev Dyn, 236:3088-99
Glycocodes
- Cadwalader EL, Condic ML, Yost HJ (2012) 2-O-sulfotransferase regulates Wnt signaling, cell adhesion and cell cycle during zebrafish epiboly. Development, 139:1296-305
- Arrington CB, Yost HJ (2009) Extra-embryonic Syndecan-2 regulates organ primordia migration and fibrillogenesis throughout the zebrafish embryo. Development, 136:3143-52
- Cadwallader AB, Yost HJ (2007) Combinatorial expression patterns of heparan sulfate sulfotransferases in zebrafish: III. 2-O-sulfotransferase and C5-epimerases. Dev Dyn, 236:581-6
- Cadwallader AB, Yost HJ (2006) Combinatorial expression patterns of heparan sulfate sulfotransferases in zebrafish: II. The 6-O-sulfotransferase family. Dev Dyn, 235:3432-7
- Cadwallader AB, Yost HJ (2006) Combinatorial expression patterns of heparan sulfate sulfotransferases in zebrafish: I. The 3-O-sulfotransferase family. Dev Dyn, 235:3423-31
