David Goldenberg
Professor of Biological Sciences, Adjunct Professor of Biochemistry, and Adjunct Professor of Chemistry
Protein Dynamics and Function, Protein Folding, Protein Biophysics

Molecular Biology Program
Biological Chemistry Program
Education
A.B. Whitman College
Ph.D. Massachusetts Institute of Technology
Research
The long-range goal of research in my laboratory is to understand the mechanisms by which polypeptide chains fold into stable structures and assemble into functional complexes. We are particularly interested in understanding the role of protein dynamics in determining the stabilities of protein structures, the specificity of their formation and their functional properties. In studying these questions, we use a variety of biophysical approaches, including NMR spectroscopy, small-angle neutron and X-ray scattering, X-ray crystallography and computational simulations.
Our current efforts are focused on two aspects of protein dynamics. The first is the characterization of unfolded proteins. Although it is usually the well-defined folded structures of proteins that are associated with biological function, there is growing interest in the properties of unfolded proteins, which are best described as broad ensembles of rapidly interconverting structures. These ensembles serve as the starting point for folding and as the reference state for most measurements of protein stability. In addition, non-native states are now known to play important roles in normal cellular processes, including protein localization and degradation, as well as pathological processes such as the formation of amyloid fibers associated with many neurodegenerative diseases. One of our major current goals is to learn how the properties of unfolded proteins change in the presence of high concentrations of other macromolecules, as found intracellularly. Using small-angle neutron scattering with solvent contrast matching, we are able to monitor the shift towards more compact conformations as the volume accessible to the unfolded molecules is reduced by molecular crowding. Our initial results have demonstrated the feasibility of this approach, and also highlight the limitations of current theoretical treatments of molecular crowding.
Another long standing interest is the interaction between a protease, trypsin, and a natural inhibitor of this enzyme, bovine pancreatic trypsin inhibitor (BPTI). BPTI is a member of a large class of natural protease inhibitors that act by binding to the active sites of their targets, much as a substrate would, but are resistant to hydrolysis. By blocking the access of potential substrates to the active sites, these inhibitors play critical biological roles in preventing unregulated proteolysis. Amino acid replacements at specific sites in BPTI can, at the same time, increase the flexibility of the free inhibitor, decrease the stability of the complex it forms with trypsin and increase its likelihood of being hydrolyzed by the enzyme. Together, these observations suggest that flexibility, or the lack thereof, is a critical factor in the function of this and other protease inhibitors.
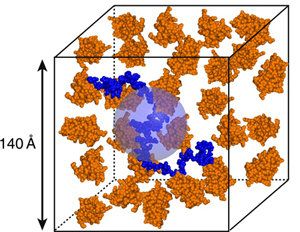
Figure caption: A scaled cartoon representation of the crowding of an intrinsically disordered protein (the bacteriophage Λ N protein, blue) by a small globular protein (orange) under the conditions used in the small-angle neutron scattering experiments described in Johansen et al., 2011.
References
- Goldenberg DP, Argyle B (2014) Minimal effects of macromolecular crowding on an intrinsically disordered protein: a small-angle neutron scattering study. Biophys J 106:905-914
- Goldenberg DP, Argyle B (2014) Self crowding of globular proteins studied by small-angle x-ray scattering. Biophys J 106:895-904
- Goldenberg DP (2013) Protein folding and assembly. In Encyclopedia of Biological Chemistry (Lennarz, W. J. & Lane, M., eds.), volume 3, pp. 625–631. Academic Press, Waltham, MA, 2nd edition
- Johansen D, Trewhella J, Goldenberg DP (2011) Fractal dimension of an intrinsically disordered protein: Small-angle X-ray scattering and computational study of the bacteriophage λ N protein. Protein Sci 20:1955-1970
- Narajan S, Amir D, Grupi A, Goldenberg DP, Minton AP, Haas E (2011) Modulation of functionally significant conformational equilibria in adenylate kinase by high concentrations of trimethylamine oxide attributed to volume exclusion. Biophys J 100:2991-2999
- Johansen D, Jeffries CMJ, Hammouda B, Trewhella J, Goldenberg DP (2011) Effects of macromolecular crowding on an intrinsically disordered protein characterized by small-angle neutron scattering with contrast matching. Biophys J 100:1120-1128
- Goldenberg DP (2010) The product operator formalism: A physical and graphical interpretation. Concepts Magn Reson Part A 36A:49-83
- Zakharova E, Horvath MP, Goldenberg DP (2009) Structure of a serine protease poised to resynthesize a peptide bond. Proc Natl Acad Sci, USA 106:11034-11039
- Zakharova E, Horvath MP, Goldenberg DP (2008) Functional and structural roles of the Cys14-Cys38 Disulfide of Bovine Pancreatic Trypsin Inhibitor. J Mol Biol 382:998-1013
- Wang Y, Trewhella J, Goldenberg DP (2008) Small-angle x-ray scattering of reduced ribonuclease A: Effects of solution conditions and comparisons with a computational model of unfolded proteins. J Mol Biol 377:1576-1592
- Hanson WM, Domek GJ, Horvath MP, Goldenberg DP (2007) Rigidification of a flexible protease inhibitor variant upon binding to trypsin. J Mol Biol 366:230-243
